In the GO, we’ve historically maintained a number of experimental extensions to the ontology as obo-format bridging files in the infamous “scratch” folder. Sometimes sets of these migrate into the main ontology (the regulation logical definitions, as well as the occurs in ones started this way). This has always been harder for axioms that point to external ontologies like chebi (although with this change it should be simpler).
These “bridging ontologies” fall into two main categories:
- logical definitions [1]
- taxon constraints[2]
As part of a general move to make more direct use of OWL in GO, we have moved the primary location of some of the bridging ontologies to the new SVN repository, with the primary format being OWL. Some of See: the Ontology extensions page on the GO wiki.
This has a number of advantages:
- the bridging axioms can be edited directly in protege, in the context of the GO and the external ontology, using an importer ontology
- we can make use of more expressive class expressions
We will provide translations back to obo but these may be lossy.
Eventually these will move into the main ontology, but there are issues regarding import chains, MIREOTing and public releases that need resolved. For now you have to explicitly import these ontologies plus any dependent ontologies. Note that this is made easier by a combination of svn:externals and catalog xml files – see tips.
Check out the repository like this:
cd svn co svn+ssh://ext.geneontology.org/share/go/svn/trunk go-trunk cd go-trunk/ontology/extensions
This should result in a directory structure like this:
ontology/
external/
cell-ontology/
ncbitaxon-slim/
..
editors/
gene_ontology_write.obo
extensions/
x-FOO.owl
x-FOO-importer.owl
...
catalog-v001.xml
The importer ontologies are designed to be loaded into Protege or programatically via OWLTools (use the new “–use-catalog” option with OWLTools).
Here is an example importer file – it contains no axioms of its own, it exists as a parent to other ontologies:
<owl:Ontology rdf:about="http://purl.obolibrary.org/obo/go/extensions/x-metazoan-anatomy-importer.owl"> <owl:imports rdf:resource="http://purl.obolibrary.org/obo/go.owl"/> <owl:imports rdf:resource="http://purl.obolibrary.org/obo/ro.owl"/> <owl:imports rdf:resource="http://purl.obolibrary.org/obo/uberon/composite-metazoan.owl"/> <owl:imports rdf:resource="http://purl.obolibrary.org/obo/go/extensions/x-cell.owl"/> <owl:imports rdf:resource="http://purl.obolibrary.org/obo/go/extensions/x-metazoan-anatomy.owl"/> </owl:Ontology>
Note that most of these will be fetched from the svn externals directory (if you load from svn, rather than the web).
After loading, you should see something like:
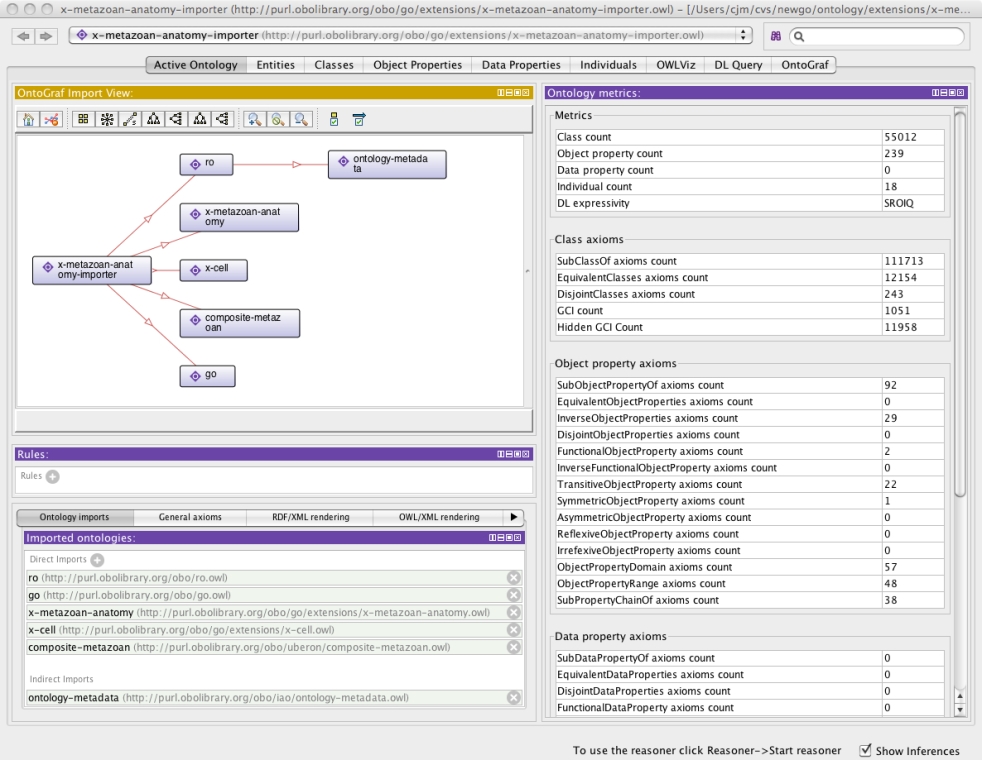
See the wiki for more details.
The bridging axioms have been under-used in the GO, expect more use of them, as part of ontology development and downstream applications over the coming year!
[1]Mungall, C. J., Bada, M., Berardini, T. Z., Deegan, J., Ireland, A., Harris, M. A., Hill, D. P., and Lomax, J. (2011). Cross-product extensions of the Gene Ontology. Journal of Biomedical Informatics 44, 80-86.PMC dx.doi.org/10.1016/j.jbi.2010.02.002
[2] Deegan Née Clark, J. I., Dimmer, E. C., and Mungall, C. J. (2010). Formalization of taxon-based constraints to detect inconsistencies in annotation and ontology development. BMC Bioinformatics 11, 530. http://www.biomedcentral.com/1471-2105/11/530