This is one post in a series of tips on ontology development, see the parent post for more details.
 A Porphyrian tree. With apologies to The Shamen, Ebeneezer Goode
A Porphyrian tree. With apologies to The Shamen, Ebeneezer Goode
The idea of classification using tree structures can be traced to the 3rd century CE and the Greek philosopher Porphyry’s Trees depicting Aristotle’s categories. The tree has enjoyed a special status despite the realization that nature can be classified along multiple axes, leading to polyhierarchies or Directed Acyclic Graphs (DAGs).
It is unfortunately still lore in some parts of that ontology community that Multiple Inheritance (MI) is bad and that Single Inheritance (SI) ontologies are somehow purer or better. This is dangerous advice, although there is a kernel of truth here. Unfortunately this kernel of truth has been misunderstood and miscommunicated, usually with bad results.
In fact, it is good ontology engineering practice to never assert MI, only infer it (see the forthcoming post on ‘Rector Normalization’). Following the Rector normalization methodology, the “primitive skeleton” should ideally form a tree, but the domain ontologies defined using these skeletons will be inferred polyhierarchies, with MI up the wazoo. This has no effect on the end-user who still consumes the ontology as a polyhierarchy, but has a huge benefit for the ontology maintainer. It should also be noted here that we are only talking about SubClass (aka is-a, aka subsumption) relationships here (see further on for notes on part-of).
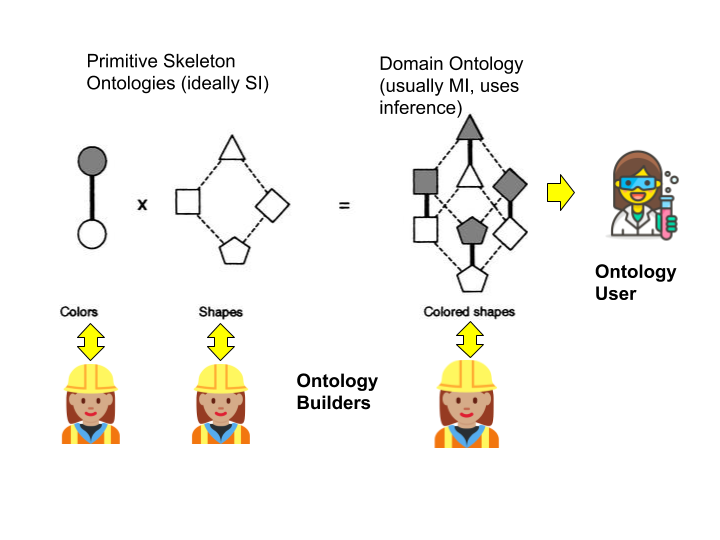 Figure: Simplified example of Rector Normalization: two primitive ontologies combined into compositional classes yielding a polyhierarchy.
Figure: Simplified example of Rector Normalization: two primitive ontologies combined into compositional classes yielding a polyhierarchy.
And additionally, it should be noted that it is also true that some ontologies do engage in IsA-overloading, which can lead to problems. The problem is that this kernel of truth has been miscommunicated, and some still cling to a purist notion of SI that is harmful. This miscommunication has resulted in ontologies that deliberately omit important links. Users are often unaware of this fact, and unaware that they are getting incomplete results.
Examples of problematic promulgations of SI
You may be reading this and are wondering why SI in ontologies would even be a thing, given that ever since the 2000 Gene Ontology paper (and probably before this) the notion of an ontology classification as a MI/DAG has been de rigueur. You may be thinking “why is he even wasting his time writing this post, it’s completely obvious”. If so, congratulations! Maybe you don’t need this article, but it may still be important for you to know this as a user, since some of the ontologies you use may have been infected with this advice. If you are part of the ontology community you have probably heard conflicting or confusing advice about SI. I hope to dispel that bad advice here. I wish I didn’t have to write this post but I have seen so much unnecessary confusion caused by this whole issue, I really want to put it to bed forever.
Here are some examples of what I consider confusing or conflicting advice:
1) This seminal paper from Schulze-Kremer and Smith has some excellent advice, but also includes the potentially dangerous:
Multiple inheritance should be carefully applied to make sure that the resulting subclasses really exist. Single inheritance is generally safer and easier to understand.
This is confusing advice. Of course, any axiom added to an ontology has to be applied carefully. It’s not clear to me that SI is easier to understand. Of course, maintaining a polyhierarchy is hard, but the advice here should be to infer the polyhierarchy.
2) The book Building Ontologies with BFO has what I consider conflicted and confusing advice. It starts well, with the recommended advice to not assert MI, but instead to infer it. It then talks about the principle of single inheritance. I hold that elevating SI to “principle” is potentially dangerous due to the likelihood of miscommunication. It lists 5 purported reasons for adhering to SI:
- computational benefits [extremely dubious, computers can obviously handle graphs fine]
- Genus-differentia definitions [I actually agree, see future post]
- enforces discipline on ontology maintainers to select the “correct” parent [dubious, and talk of “enforcing discipline” is a red flag]
- ease of combining ontologies [very dubious]
- users find it easier to find the terms they require using an “official” SI monohierarchical version of the ontology [dubious/wrong. this confuses a UI issue with an ontology principle, and conflicts with existing practice].
3) The Foundational Model of Anatomy (FMA) is a venerable ontology in the life sciences, it’s FAQ contains a very direct statement advocating SI:
2) Why do the FMA authors use single inheritance?
The authors believe that single inheritance assures the true essence of a class on a given context.
I don’t understand what this means, and as I show in the example below, this adherence to the SI is to the detriment of users of the FMA.
4) The disease ontology HumanDO.obo file is single-inheritance, as is the official DO browser.
doid-non-classified.obo : DO’s single asserted is_a hierarchy (HumanDO.obo), this file does not contain logical definitions

Figure: official browser for DO is SI: lung carcinoma is classified as lung cancer, but no parentage to carcinoma. Users querying the SI version for carcinoma would not get genes associated with lung carcinoma. Most other browsers such as OLS and the Alliance disease pages show the MI version of DO, where this class correctly has two is-a parents.
The SI principle leads to massively incomplete query results; an example from the FMA
I will choose the FMA as an example of why the SI principle in its strict form is dangerous. The following figure shows the classification of proximal phalanx of middle finger (PPoMF) in FMA. The FMA’s provided is-a links are shown as black arrows. I have indicated the missing is-a relationship with a dashed red line.

The FMA is missing a relationship here. Of course, many ontologies have missing relationships, but in this case the missing relationship is by design. If the FMA were to add this relationship it would be in violation of one of its stated core principles. In elevating this purist SI principle, the FMA is less useful to users. For example, if I want to classify skeletal phenotypes, and I have a phenotype involving the proximal phalanx of middle finger (PPoMF), and a user queries for proximal phalanx of [any] finger (PPoF), the user will not get the expected results. Unfortunately, there many cases of this in the FMA, since so many of the 70k+ classes are compositional in nature, and many FMA users may be unaware of these missing relationships. One of the fundamental use cases of ontologies is to be able to summarize data at different levels, and to discard this in the name of purity seems to me to be fundamentally wrongheaded. It is a simple mathematical fact that when you have compositional classes in an ontology, you logically have multiple inheritance.
This exemplifies a situation where SI is the most dangerous, when the following factors occur together: (1) the ontology is SI (2) the ontology has many compositional classes (there are over 70k classes in the FMA) (3) there are no logical definitions / Rector normalization, hence no hope of inferring the complete classification (4) The ontology is widely used.
The FMA is not the only ontology to do this. Others do this, or have confusing or contradictory principles around SI. I have chosen to highlight the FMA as it is a long established ontology, it is influential and often held up as an exemplar ontology by formal ontologists, and it is unambiguous in its promulgation of SI as a principle. It should be stressed that FMA is excellent in other regards, but is let down by allowing dubious philosophy to trump utility.
The advice we should be giving
The single most important advice we should be giving is that ontologies must be useful for users, and they must be complete, and correct. Any other philosophical or engineering principle or guideline that interferes with this must be thrown out.
We should also be giving advice on how to build ontologies that are easy to maintain, and have good practice to ensure completeness and correctness. One aspect of this is that asserting multiple is-a parents is to be avoided, these should be inferred. In fact this advice is largely subsumed within any kind of tutorial on building OWL ontologies using reasoning. Given this, ontologists should stop making pronouncements on SI or MI at all, as this is prone to misinterpretation. Instead the emphasis for ontology developers should be on good engineering practice.
TL;DR
multiple inheritance is fine. If you’re building ontologies add logical definitions to compositional classes, and use reasoning to infer superclasses.
Hello Chris,
I find your blog very useful.
I have a question on this post, what is IsA-overloading and what problems it may cause?
Hi Citlalli – this is probably deserving of it’s own post! But briefly, this is when isA (subClassOf between two named classes) is used where a different relationship is more appropriate. For example, if we say finger isA hand. But there are more nuanced cases. Even when valid it can make the ontology harder to maintain
Oh, I get the idea of what it is. If you decide to write a post for the subject, I will readily read it. Thank you for your reply.
Another nice blog post, thanks for writing it!
I feel that for biologists outside the ontology community, SI hierarchies are way more widely known. Maybe because species taxonomy is currently modeled largely as tree, based on inferred evolutionary similarity.
For cell types, I see many articles trying to fit cell types to a tree structure, which would strictly entail SI (ex: https://bmcgenomics.biomedcentral.com/articles/10.1186/s12864-015-2297-3) which seems a bit too constrained. I guess it is like the saying, “if all you have is a hammer, everything looks like a nail” (https://en.wikipedia.org/wiki/Law_of_the_instrument).
An analogous hammer-nail situation may happen to people trying to fit cell types into periodic tables (https://dev.biologists.org/content/146/12/dev169854), which feels quite unnatural to me, as I do not see any periodicity.
Anyways, do you any see any cases of multiple inheritances in “mainstream” biological classification, of the kind that would be present in a high-school biology course, for example?
I wasn’t aware of the periodic table paper, interesting, thanks!
In my experience biologists don’t have such a problem with MI, the objection to it is coming from the philosophical camp. However, I can the way things are going in clustering scSeq results I see a lot of dendrograms…