This article is part of the OntoTips series.
A common structure found in many ontologies is the single child pattern. I consider this an anti-pattern, to be avoided.
The most common form is with is_a children (i.e subClassOf between two named classes), but the anti-pattern also applies to other relationship types. We can formalize the single child subclass pattern as:
- C1 direct SubClassOf P
- NOT exists some C2, such as C2 is a direct SubClassof P, and C2 != C1
Depicted as:

One reason this is an anti-pattern is that it is inherently incomplete. i.e. there must be instances of P that are not instances of C1 (otherwise why have two classes – see the reflexive subclass anti-pattern). Following a principle of reasonable completeness (see open world post) we should include sibling terms where appropriate.
Here is a concrete example from a fictional ontology:

Here there is a single specialization of a disease term, based on severity.
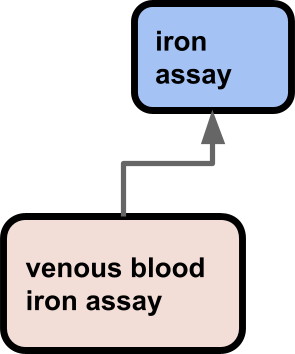
Another example (adapted from an existing ontology):

Here there is a specialization of the assay term based on a property of the pool or iron.
A different example (adapted from an existing ontology):
This kind of structure is not uncommon in many OBO ontologies. And there is a reasonable defense: we have limited ontology editing resources, and many terms are added on request. Curators are free to request a more specific term if they feel it is necessary for annotating (e.g a disease that has as phenotype mild flu) but they may have no need for the implicit sibling terms. And ontology developers see no need to do additional work they are not requested to do.
However, this leads to lopsided ontologies that are often confusing for people not deeply immersed the development of these ontologies. It is hard to tell if omissions are intentional or unintentional. And the practice of instantiating single children has bad downstream effects of annotation, this is something we have frequently observed over multiple ontologies.
Consider the flu example above. A new annotator may want to annotate a disease that has a severe flu phenotype. They may make an implicit assumption that choosing the parent term ‘flu’ would communicate ‘severe flu’; if it was mild, they would have selected ‘mild flu’. But this is not the explicit assertion they are making – they are making a closed world assumption that doesn’t hold for the logic of the ontology. While some of this can be obviated with training, and ensuring curators request specific sibling terms rather than trying to let the parent do the work. But many single-child cases are in fact more nuanced that the flu example.
Instead, it is better to take a more prospective approach to ontology development, try and anticipate in advance terms that may be required, and populate them in a balanced fashion – this will result in more balanced annotations. It is much easier to do this if you follow OWL axiomatization and have a formal design pattern system such as DOSDPs. In fact you can use such a system to automate detection of single-child patterns and imbalances.
While it is trivial to detect single is-a children using a SPARQL query encoding the pattern above, it won’t capture the more nuanced cases of single children by a given axis of classification.
Consider this made-up ontology structure, where we have a parent class with only two subclasses explicitly populated:

In this particular example, both are also single-children via an axis of classification. While on a gross structural level the lower terms each has a sibling, each sibling is clearly classified differently. The first is classified along classical taxonomic/evolutionary descent terms, the second is by a different property.
The above example is made-up and would strike most people as bad design (even if strictly logically coherent). Where is the concept of inedible animals, where are the invertebrates (and indeed edible and inedible vertebrates and invertebrates)?
But in fact this antipattern plagues most OBO ontologies. These are harder to spot, especially if the ontology is unaxiomatized.
For example:

Structurally this doesn’t look like a single-child anti-pattern, but it is in fact an example of a single-child-by-axis pattern. And if there are no subclasses, this is an instance of the ragged lattice pattern, which I will cover in a future post.
While these can’t be detected by straightforward SPARQL queries, if you use a system such as DOSDPs you can use this to analyze your ontology for these structures, and proactively guard against them.
While the above examples all focus on subClassOf/is-a relations, the same guidelines apply to other edge labels. For example, if an anatomy ontology only listed a single part of the head (such as the mouth):

Most people would consider this poor design. While of course it’s true it’s unreasonable to expect ontologies to be complete, the reasonable completeness principle should apply, and if for some reason this not unattainable, at the very least the incompleteness needs to be clearly documented.
In closing, as ontology developers it can be tempting to ignore these single child cases – we have limited resources, and must balance this with being able to provide users with terms they request, which may lead to spottiness and incompleteness. But ignoring these just leads to more work downstream, and in some cases it can lead to incomplete annotation. So avoid single is-a children!
Chris, while I subscribe to most of what you write, I believe there is at least one situation when a single-child pattern is pretty sane. Namely, when you have natural subclasses such that one is very common (for example, 50% of the superclass’ instances fall under it) and the remaining subclasses are numerous but each of them is rare alone (for example, up to 5%). Then by only having the common subclass present in the ontology you balance well the discriminativeness and parsimony. Since I believe that sibling classes such as OtherX are an even clearer anti-pattern (both semantically and in terms of maintenance).
I would also say that some of the examples you showed might also be candidates for taxonomy disentangling using A. Rector’s normalization pattern after all…
Thanks Vojtěch. I agree that OtherX is an anti-pattern, but ComplementOfX need not be (especially if there is a natural name for the complement). We commonly apply this in Mondo, where we have mendelian and non-mendelian subtypes of a disease. But often in these cases you have more specific subclasses anyway. Are there some concrete examples you are thinking of? The most common case of justifiable single-children to me are in organism taxonomies translated to ontologies, where there are often long linear chains of single children, but this is expected given the need to balance phylogeny and the need to populate taxa at different levels.
Yes, organism taxonomies is what I had primarily in mind. Though by intuition rather than by experience with real biomed projects. My own ontology projects are rather on linked data schemas – where parsimony matters even more. In the LD world the classes are primarily considered in connection with sets of their instances rather than in the context of their ontology graph, so the ¨aesthetics¨ of the taxonomy is secondary.