A frequent source of confusion with ontologies and more generally with any kind of information system is the Open World Assumption. This trips up novice inexperienced users, but as I will argue in this post, information providers could do much more to help these users. But first an explanation of the terms:
With the Open World Assumption (OWA) we do not make any assumptions based on the absence of statements. In contrast, with the Closed World Assumption (CWA), if something is not explicitly stated to be true, it is assumed to be false. As an example, consider a pet-owner database with the following facts:
Fred type Human .
Farrah type Human .
Foofoo type Cat .
Fido type Dog .
Fred owns Foofoo .
Farrah owns Fido.
Depicted as:

Under the CWA, the answer to the question “how many cats does Fred own” is 1. Similarly, for “how many owners does Fido have” the answer also 1.
RDF and OWL are built on the OWA, where the answer to both question is: at least 1. We can’t rule out that Fred also owns Fido, or that he owns animals not known to the database. With the OWA, we can answer the question “does Fred own Foofoo” decisively with a “yes”, but if we ask “does Fred own Fido” the answer is “we don’t know”. It’s not asserted or entailed in the database, and neither is the negation.
Ontology formalisms such as OWL are explicitly built on the OWA, whereas traditional database systems have features constructed on the CWA.
OWL gives you mechanisms to add closure axioms, which allows you to be precise about what is known not be to true, in addition what is known to be true. For example, we can state that Fred does not own Fido, which closes the world a little. We can also state that Fred only owns Cats, which closes the world further, but still does not rule out that Fred owns cats other than Foofoo. We can also use an OWL Enumeration construct to exhaustively list animals Fred does own, which finally allows the answer to the question “how many animals does Fred own” with a specific number.
OWL ontologies and databases (aka ABoxes) often lack sufficient closure axioms in order to answer questions involving negation or counts. Sometimes this is simply because it’s a lot of work to add these additional axioms, work that doesn’t always have a payoff given typical OWL use cases. But often it is because of a mismatch between what the database/ontology author thinks they are saying, and what they are actually saying under the OWA. This kind of mismatch intent is quite common with OWL ontology developers.
Another common trap is reading OWL axioms such as Domain and Range as Closed World constraints, as they might be applied in a traditional database or a CWA object-oriented formalism such as UML.
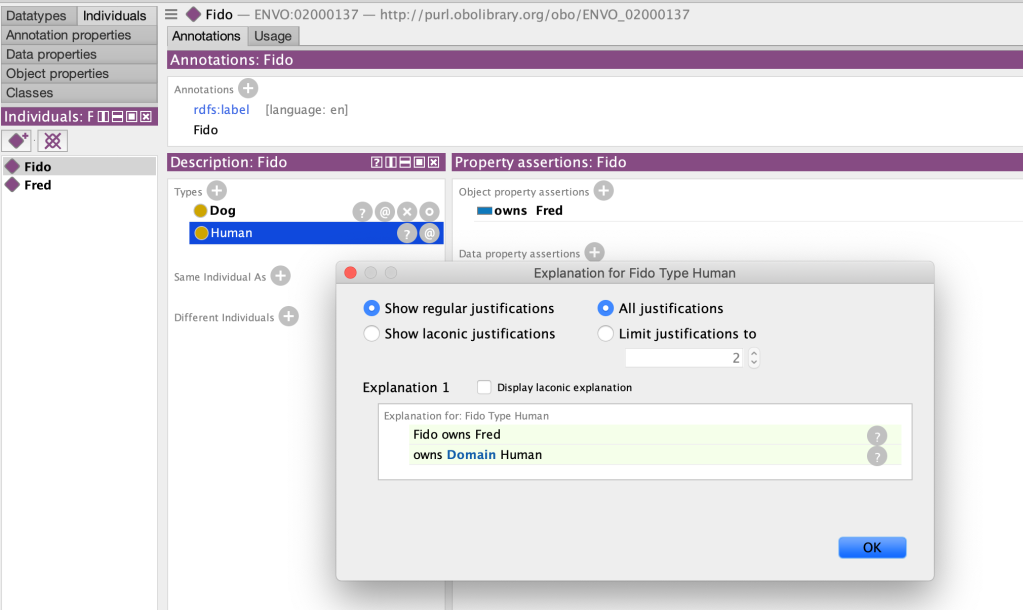
Consider the following database plus ontology in OWL, where we attempt to constrain the ‘owns’ property only to humans
owns Domain Human
Fido type Dog
Fred type Human
Fido owns Fred
We might expect this to yield some kind of error. Clearly using our own knowledge of the world something is amiss here (likely the directions of the final triple has been accidentally inverted). But if we are to feed this in to an OWL reasoner to check for incoherencies (see previous posts on this topic), then it will report everything as consistent. However, if we examine the inferences closely, we will see that it is has inferred Fido to be both a Dog and a Human. It is only after we have stated explicit axioms that assert or entail Dog and Human are disjoint that we will see an inconsistency:
In many cases the OWA is the most appropriate formalism to use, especially in a domain such as the biosciences, where knowledge (and consequently our databases) is frequently incomplete. However, we can’t afford to ignore the fact that the OWA contradicts many user expectations about information systems, and must be pragmatic and take care not to lead users astray.
BioPAX and the Open World Assumption
BioPAX is an RDF-based format for exchanging pathways. It is supposedly an RDF/OWL-based standard, with an OWL ontology defining the various classes and properties that can be used in the RDF representation. However, by their own admission the authors of the format were not aware of OWL semantics, and the OWA specifically, as explained in the official docs in the level 2 doc appendix, and also further expanded on in a paper from 2005 by Alan Ruttenberg, Jonathan Rees, and Joanne Luciano, Experience Using OWL DL for the Exchange of Biological Pathway Information, in particular the section “Ambushed by the Open World Assumption“. This gives particular examples of where the OWA makes things hard that should be easy, such as enumerating the members of a protein complex (we typically know all the members, but the BioPAX RDF representation doesn’t close the world).

The Ortholog Conjecture and the Open World Assumption

In 2011 Nehrt et al made the controversial claim that they had overturned the ortholog conjecture, i.e they claimed that orthologs were less functionally similar than paralogs. This was in contrast to the received wisdom, i.e if a gene duplicates with a species lineage (paralogs) there is redundancy and one copy is less constrained to evolve a new function. Their analysis was based on semantic similarity of annotations in the Gene Ontology.
The paper stimulated a lot of discussion and follow-up studies and analyses. We in the Gene Ontology Consortium published a short response, “On the Use of Gene Ontology Annotations to Assess Functional Similarity among Orthologs and Paralogs: A Short Report“. In this we pointed out that the analysis assumed the CWA (absence of function assignment means the gene does not have that function), whereas GO annotations should be interpreted under the OWA (we have an explicit way of assigning that a gene does not have a function, rather than relying on absence). Due to bias in GO annotations, paralogs may artificially have higher functional similarity scores, rendering the original analysis insufficient to reject the ortholog conjecture.
The OWA in GO annotations is also addressed in the GO Handbook in the chapter Pitfalls, Biases, and Remedies by Pascale Gaudet. This chapter also makes the point that OWA can be considered in the context of annotation bias. For example, not all genes are annotated at the same level of specificity. The genes that are annotated reflect biases in experiments and publication, as well as what is selected to be curated.
Open World Assumption Considered (Sometimes) Harmful
The OWA is harmful where it grossly misaligns with use expectations.
While a base assumption of OWA is usually required with any curated information, it is also helpful to think in terms of an overriding implicit contract between any information provider and information consumer: any (good) information provider attempts to provide as complete information as is possible, given resource constraints.
My squid has no tentacles
Let’s take an example: If I am providing an ontology I purport to be an anatomical ontology for squid, then it behooves me to make sure the main body parts found in a squid are present.

Let’s say my ontology contains classes for some squid body parts such as eye, brain, yet lacks classes for others such as the tentacle. A user may be surprised and disappointed when they search for tentacle and come back empty-handed (or empty tentacled, if they are a squid user). If this user were to tell me that my ontology sucked, I would be perfectly within my logical rights to retort: “sir, this ontology is in OWL and thus follows the Open World Assumption; as such the absence of a tentacle class in my squid ontology does not entail that squids lack tentacles, for such a claim would be ridiculous. Please refer to this dense interlinked set of documents published by the W3C that requires PhD in logic to understand and cease from making unwarranted assumptions about my ontology“.
Yet really the user is correct here. There should be an assumption of reasonable coverage, and I have violated that assumption. The tentacle is a major body part, it’s not like I have omitted some obscure neuroanatomical region. Is there a hard and fast dividing line here? No, of course not. But there are basic common sense principles that should be adhered to, and if they cannot be adhered to, omissions and biases should be clearly documented in the ontology to avoid confusing users.
This hypothetical example is made up, but I have seen many cases where biases and omissions in ontologies confusingly lead the user to infer absence where the inference is unwarranted.
Hydroxycholoroquine and COVID-19
The Coronavirus Infectious Disease Ontology (CIDO) integrates a number of different ontologies and includes axioms connecting terms or entities using different object properties. An example is the ‘treatment-for’ edge which connects diseases to treatments. Initially the ontology only contained a single treatment axiom, between COVID-19 and Hydroxychloroquine (HCQ). Under the OWA, this is perfectly valid: COVID-19 has been treated with HCQ (there is no implication about whether treatment is successful or not). However, the inclusion of a single edge of this type is at best confusing. A user could be led to believe there was something special about HCQ compared to other treatments, and the ontology developers had deliberately omitted these. In fact initial evidence for HCQ as a successful treatment has not panned out (despite what some prominent adherents may say). There are many other treatments, many of which are in different clinical trial phases, many of which may prove more effective, yet assertions about these are lacking in CIDO. In this particular case, even though the OWA allows us to legitimately omit information, from a common sense perspective, less is more here: it is better to include no information about treatments at all rather than confusingly sparse information. Luckily the CIDO developers have rapidly addressed this situation.
Ragged Lattices, including species-specific classes
An under-appreciated problem is the confusion ragged ontology lattices can cause users. This can be seen as a mismatch between localized CWA expectations on the part of the user and OWA on the part of the ontology provider. But first an explanation of what I mean by ragged lattice:
Many ontologies are compositional in nature. In a previous post we discussed how the Rector Normalization pattern could be used to automate classification. The resulting multi-parent classification forms a lattice. I have also written about how we should embrace multiple inheritance. One caveat to both of these pieces is that we should be aware of the confusion that can be caused by inconsistently populated (‘ragged’) lattices.
Take for example cell types, which can be classified along a number of orthogonal axes, many intrinsic to the cell itself – its morphological properties, it’s lineage, its function, or gene products expressed. The example below shows the leukocyte hierarchy in CL, largely based on intrinsic properties:
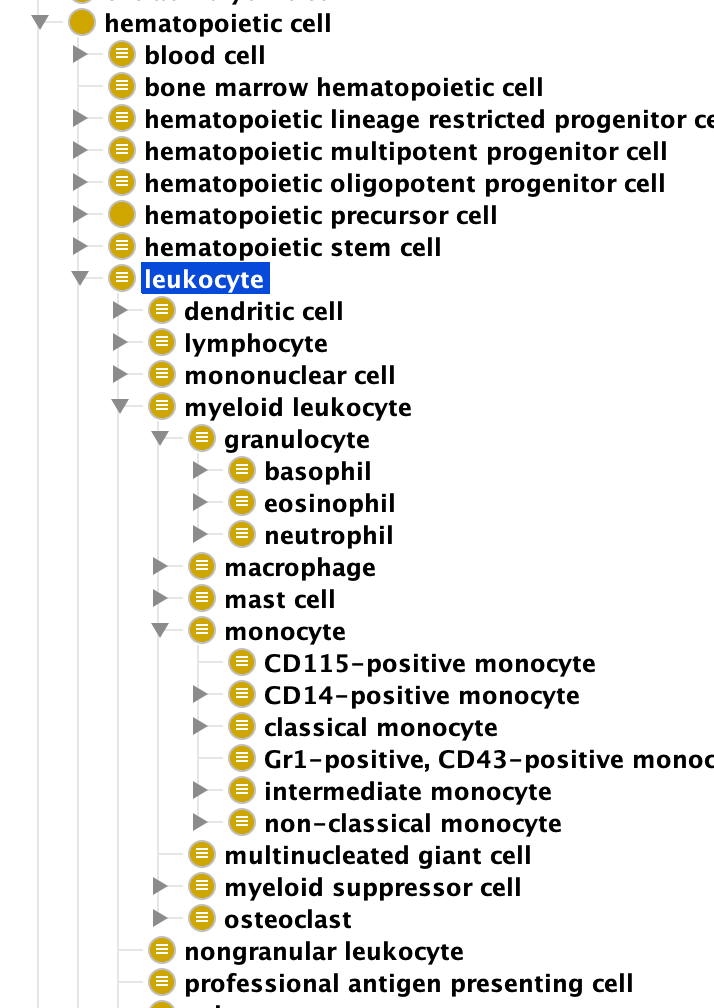
Another way to classify cells is by anatomical location. In CL we have a class ‘kidney cell’ which is logically defined as ‘any cell that is part of a kidney’. This branch of CL recapitulates anatomy at the upper levels.

so far, perfectly coherent. However, the resulting structure can be confusing to someone now used to thinking in OWL and the OWA. I have seen many instances where a user will go to a branch of CL such as ‘kidney cell‘ and start looking for a class such as ‘mast cell‘. It’s perfectly reasonable for them to look here, as mast cells are found in most organs. However, CL does not place ‘mast cell’ as a subclass of ‘kidney cell’ as this would entail that all mast cells are found in the kidney. And, CL has not populated the cross-product of all the main immune cell types with the anatomical structures in which they can be found. The fleshing out of the lattice is inconsistent, leading to confusion caused by violation of an assumed contract (provision of a class “kidney cell” and incomplete cell types underneath).
This is even more apparent if we introduce different axes of classification, such as the organism taxon in which the cell type is found, e.g. “mouse lymphocyte”, “human lymphocyte”:

Above is a screenshot of what happens when we introduce classes such as ‘mouse cell’ or ‘mouse lymphocyte’. We see very few classes underneath. Many people indoctrinated/experienced with OWL will not have a problem with this, they understand that these groupings are just for mouse-specific classes, and that the OWA holds, and absence of a particular compositional class, e.g. “mouse neuron” does not entail that mice do not have neurons.
One ontology in which the taxon pattern does work is the protein ontology, which includes groupings like “mouse protein”. PRO includes all known mouse proteins under this, so the classification is not ragged in the same way as the examples above.
There is no perfect solution here. Enforcing single inheritance does not work. Compositional class groupings are useful. However, ontology developers should try and avoid ragged lattices, and where possible populate lattices consistently. We need better tools here, e.g. ways to quantitative measure the raggedness of our ontologies.
Ontologies and databases should better document biases and assumptions
As providers of information, we need to do a better job of making all assumptions explicit and well-documented. This applies particularly to any curated corpus of knowledge, but in particular to ontologies. Even though hiding behind the OWA is logically defensible, we need to make things more intuitive for users.
It’s not uncommon for an ontology to have excellent very complete coverage of one aspect of the domain, and to be highly incomplete in another (reflecting either the biases/interests of the developers, or of the broader field). In fact I have been guilty of this in ontologies I have built or contributed to. I have seen users become confused when a class they expected to find was not present, or they have been perplexed by the ragged lattice problem, or an edge they expected to find was not present.
Few knowledge bases can ever be complete, but we can do better at documenting known unknowns or incompletenesses. We can imagine a range of formal computable ways of doing this, but a good start would be some simple standard annotation properties that can be used as inline documentation in the ontology. Branches of the ontology could be tagged in this way, e.g. to indicate that ‘kidney cell’ doesn’t include all cells found in the kidney, only kidney specific ones; or that developmental processes in GO are biased towards human and model organisms. This system could also be used for Knowledge Graphs and annotation databases too, to indicate that particular genes may be under-studied or under-annotated, an extension of the ND evidence type used in GO.
In addition we could do a better job at providing consistent levels of coverage of annotations or classes. There are tradeoffs here, as we naturally do not want to omit anything, but we can do a better job at balancing things out. Better tools are required here for detecting imbalances and helping populate information in a more balanced consistent fashion. Some of these may already exist and I’m not aware of them – please respond in the comments if you are aware of any!
Your text (really good, by the way) got me thinking about enrichment analysis, widely used in modern bioinformatics (e.g. GSEA: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1239896/). It definitely operates under the CWA for curated gene sets, at least for the calculation of the p-values when using the Gene Ontology.
In practice, I`d guess people see it as a kind of assumption of reasonable coverage, acknowledging the fact that our knowledge may change in the future. Given that the GSEA article has been cited over 23k times, I wonder if there are any practical implications…
Thanks! Yes, I think enrichment works best with reasonable coverage. I’m not sure what the practical implications of this are. I don’t think it leads to enrichment results being wrong. But the absence of enrichment may falsely lead a researcher to believe there is no patterns in their data. It may be interesting to do a tool that is probabilistic and follows the OWA, with priors for missing annotations. This would be best done with model-based rather than frequentist enrichment frameworks, e.g. https://pubmed.ncbi.nlm.nih.gov/20172960/